Arabidopsis Thaliana Root Cell Segmentation Challenge
About
The work flow of cell biologists depends more and more on a large
number of images collected from their experiments. These images
have to be analyzed, and both the inherent time loss and the human
subjectivity motivate the automation of such processes. One of
the most important steps in automatic cell biology image analysis is
the detection or segmentation of the objects (cells or other agents)
of interest.
Biology cell images resulting from confocal microscopy, where fluorescent
proteins are used, suffer from problems which make the segmentation
task more challenging:
- High Poison image noise
- Low contrast between objects and background
- Multi-spectral cross-bleed from different fluorescence dies.
The images in this challenge are resulting from in vivo plant
imaging and contain both one channel with cell wall fluorescence
information and another channel with nuclei fluorescence information.
The images were annotated by a biologist with the location
of cell walls and nucleus when visible.
The main objective of this challenge is to provide a common
basis for comparison of diverse methodologies in order to
bring greater interest to this important area of research and to
provide a way to validate possible approaches for this problem.
Task and evaluation
The task is to automatically obtain the information on the location of the cell wall and nucleus in the root image from the original multi-spectral information. The annotated data is in the form of two different image each containing non-connected components corresponding to nuclei and cell walls in the image, as this example demonstrates:


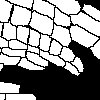
-- Original Image - Annotated Nuclei - Annotated Cell wall --
The evaluation is based on both hit/miss precision recall of the detection of each cell's wall and nucleus, as well as the coverage of the automatic segmentation area with relation to the ground truth. The main evaluation will be based on Dice's Coeficient:
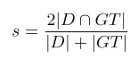
where D is a certain detection and GT is a certain annotated groundtruth (applicable
to both nuclei and cell walls), and D ∩ GT is the area intercection and D + GT is the area union.
If the Dice's coeficient between detection and annotated groundtruth is above 0.5 a True Positice (TP)
is considered to be found. All detection with have no corresponding groundtruth are False Positives (FP)
and all annotated groundtruth with no corresponding detection is a False Negative (FN).
For complete evaluation the average per image of FP, TP an FN mus be presented. Additionally the average
Dice's coeficient for all TP must be calculated and presented.
Publications can be presented on only cell wall or cell nuclei detection or both.
Publishing
All work performed in the task of cell nuclei and cell wall detection must be submitted for publication within the rules of the ICIAR 2010 conference. Publication will not depend on the cell detection performance of each method but on the scientific value of the proposed approach.
All the data used in this challenge was gathered and annotated within the scope of the European project: "Integrated Analysis of Stem Cell Function In Plant Growth and Development", part of the European Network in Plant Genomics (ERA-NET PG). Performed in Portugal under FCT contract ERAPG/0007/2006
All images have been collected at the Department of Biology of the Utrecht University by the Ben Scheres Lab.